发布者:Primerbank 时间:2017-09-22 浏览量:938
The aim of this study was to describe the first development and evaluation of a multiplex tandem PCR (MT-PCR) assay for the detection and identification of 4 common pathogenic protozoan parasites, Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis, from human clinical samples. A total of 472 fecal samples submitted to the Department of Microbiology at St. Vincent's Hospital were included in the study. The MT-PCR assay was compared to four real-time PCR (RT-PCR) assays and microscopy by a traditional modified iron hematoxylin stain. The MT-PCR detected 28 G. intestinalis, 26 D. fragilis, 11 E. histolytica, and 9 Cryptosporidium sp. isolates. Detection and identification of the fecal protozoa by MT-PCR demonstrated 100% correlation with the RT-PCR results, and compared to RT-PCR, MT-PCR exhibited 100% sensitivity and specificity, while traditional microscopy of stained fixed fecal smears exhibited sensitivities and specificities of 56% and 100% for Cryptosporidium spp., 38% and 99% for D. fragilis, 47% and 97% for E. histolytica, and 50% and 100% for G. intestinalis. No cross-reactivity was detected in 100 stool samples containing various other bacterial, viral, and protozoan species. The MT-PCR assay was able to provide rapid, sensitive, and specific simultaneous detection and identification of the four most important diarrhea-causing protozoan parasites that infect humans. This study also highlights the lack of sensitivity demonstrated by microscopy, and thus, molecular methods such as MT-PCR must be considered the diagnostic methods of choice for enteric protozoan parasites.
Enteric protozoa continue to be the most commonly encountered parasitic diseases and to cause significant morbidity and mortality throughout both developed and developing regions of the world, affecting millions of people each year (22). While Blastocystis hominis is the most common protozoan parasite detected in stool samples, the major etiological agents of parasitic diarrhea are considered to be Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis (22).
E. histolytica is a pathogenic amoeboid protozoan parasite for which humans are the primary reservoir (27). It is a potentially invasive pathogen and the causative agent of amebiasis, with approximately 50 million cases and 100,000 deaths annually (28). The clinical presentation can range from asymptomatic carriage to gastrointestinal disease and invasive disease. E. histolytica is morphologically identical to the nonpathogenic species Entamoeba dispar and Entamoeba moshkovskii, though genetic differences have confirmed their separation into independent species (4, 20). Due to this conserved morphology, stained smears of stool specimens are insufficient for differentiation of the species. Currently, the only way to differentiate the pathogenic E. histolytica from the nonpathogenic strains via microscopy is to visualize ingested red blood cells within the E. histolytica trophozoite; however, this is rarely seen in clinical samples containing E. histolytica and thus cannot be solely relied upon for diagnostic confirmation. Several studies have shown that the nonpathogenic E. dispar is more frequently encountered in clinical samples than E. histolytica, and E. moshkovskii has recently been identified from human samples from various parts of the world (5-7, 25). While staining of fixed fecal smears can determine the presence of the E. histolytica-E. dispar-E. moshkovskii complex within a stool specimen, other techniques, such as PCR or enzyme-linked immunosorbent assay (ELISA), must be employed for differentiation of the species. A number of PCR assays are available for detection and/or differentiation of Entamoeba species and have become the methods of choice due to excellent sensitivity and specificity (3, 11, 19).
G. intestinalis is a common and ubiquitous flagellate parasite with a worldwide distribution and is considered one of the main nonviral causes of diarrhea in industrialized countries (22). Clinical presentation ranges from asymptomatic carriage to acute and chronic gastrointestinal infections. The acute phase is often followed by a subacute or chronic phase of infection (22). Various diagnostic modalities are available for the detection of Giardia in clinical samples. Giardia infection can be diagnosed microscopically by identification of cysts and trophozoites in stained or unstained fecal smears. Enzyme immunoassays and immunochromatographic and direct fluorescence assays for detection of G. intestinalis in stool have been available in the form of commercial kits for several years (31). These kits are commonly used in diagnostic laboratories. A number of PCR assays are also available for the detection ofGiardia in stool specimens and show excellent sensitivity and specificity compared to microscopy and antigen detection methods (8, 10, 30).
Cryptosporidium species have a worldwide distribution and the ability to infect a wide range of vertebrate hosts (17). Cryptosporidium parvum and Cryptosporidium hominis are the species most commonly associated with human cryptosporidiosis (13), though infections with other species, such as Cryptosporidium felis and Cryptosporidium meleagridis (18), have been reported (13, 15). Infection is acquired via the fecal-oral route, and C. parvum has been recognized as the cause of large waterborne and food-borne outbreaks of gastroenteritis. Patients tend to present with a self-limiting diarrhea that may last for several weeks to months, with the highest burden of disease occurring in children under 5 years of age (1). In immunosuppressed patients, the disease is often more severe, is usually associated with chronic diarrhea and wasting, and can be life threatening (1). Laboratory diagnosis of cryptosporidiosis traditionally relies on special staining techniques, while alternative diagnostic techniques have also been employed, including ELISA and various PCR assays (2, 9, 12, 32). A number of studies have demonstrated that PCR has superior sensitivity for the detection of Cryptosporidium species compared to conventional staining, microscopy, and ELISA (14, 16).
D. fragilis is a pathogenic parasite with a worldwide distribution that has been shown to cause acute gastrointestinal disease, with chronic infections also documented (26). The most frequent clinical symptoms associated with D. fragilis are diarrhea and abdominal pain (26). Traditional diagnosis of D. fragilis is based on prompt fixation and permanent staining, as trophozoites degenerate within hours of being passed, with demonstration of the characteristic nuclear structure achieved by permanent stained preparations only. Such techniques are time-consuming and require experienced personnel to interpret the stained smears. Newer molecular techniques, conventional and real-time PCR (RT-PCR) targeting the small-subunit (SSU) ribosomal DNA (rDNA), have been developed for the diagnosis of D. fragilis and offer greater sensitivity and specificity than microscopy (23, 24).
In summary, this is the first study to develop and evaluate a multiplex tandem PCR (MT-PCR) assay for the simultaneous detection and identification of Cryptosporidium, D. fragilis, E. histolytica, and Giardia in human fecal samples. Additionally, an internal control for the detection of inhibition of the amplification by fecal contaminants was included in the assay. Traditionally, microscopy has been the method of choice; however, for diagnosis of enteric protozoans, molecular methods are now considered the gold standard for diagnosis, given the excellent sensitivities and specificities achieved by molecular methods. Although PCR-based assays have been successfully used for all four organisms, this is the first assay to provide detection of the four different targets in one commercially available kit. The performance of the assay was evaluated with a range of clinical samples and controls and compared to both RT-PCR and conventional microscopy.
Fecal specimens.Fecal specimens (n = 472) submitted to the Department of Microbiology at St. Vincent's Hospital, Sydney, Australia, for investigation of diarrhea from March 2009 until March 2010 were included in the study. Specimens from outpatients were collected by the patient and submitted to the laboratory as a fresh specimen, along with a portion mixed with sodium acetate-acetic acid-formalin (SAF) preservative. Specimens from inpatients or specimens received without a portion fixed in SAF were immediately preserved in SAF upon arrival at the laboratory (approximately 1 g of feces was placed into SAF and fixed overnight). All stool samples were subjected to microscopy, RT-PCR, and MT-PCR as described below.
Microscopy.The SAF-fixed specimens were stained using a modified iron hematoxylin stain incorporating a carbol fuchsin step to detect coccidia (Fronine, Australia) according to the manufacturer's recommendations and examined by oil immersion microscopy (×1,000 magnification). Approximately 250 fields of view were examined on each slide. All slides were stored for the duration of the project and reviewed if discrepant results were obtained with the PCR assays.
DNA extraction.All unfixed stool samples (n = 472) underwent direct DNA extraction using the QIAamp DNA stool minikit (Qiagen, Hilden, Germany) using a portion of the fresh stool sample as previously described (23).
RT-PCR.Cryptosporidium spp., E. histolytica, and G. intestinalis PCRs were performed on all samples as previously described using the following primers and probes: E. histolytica, Ehd-239F (5′-ATTGTCGTGGCATCCTAACTCA-3′), Ehd-88R (5′-GCGGACGGCTCATTATAACA-3′), and histolytica-96T (VIC-5′-TCATTGAATGAATTGGCCATTT-3′-nonfluorescent quencher); G. intestinalis, Giardia-80F (5′-GACGGCTCAGGACAACGGTT-3′), Giardia-127R (5′-TTGCCAGCGGTGTCCG-3′), and Giardia-105T (6-carboxyfluorescein [FAM]-5′-CCCGCGGCGGTCCCTGCTAG-3′-black hole quencher 1); Cryptosporidiumspp., CrF (5′-CGCTTCTCTAGCCTTTCATGA-3′), CrR (5′-CTTCACGTGTGTTTGCCAAT-3′), and Crypto (Texas Red-5′-CCAATCACAGAATCATCAGAATCGACTGGTATC-3′-black hole quencher 2) (29). D. fragilis RT-PCR was performed on all samples, amplifying a 77-bp SSU rDNA product as previously described (21), using the primers DF3 (5′-GTTGAATACGTCCCTGCCCTTT-3′) and DF4 (5′-TGATCCAATGATTTCACCGAGTCA-3′) and TaqMan probe (FAM-5′-CACACCGCCCGTCGCTCCTACCG-6-3′- 6-carboxytetramethylrhodamine [TAMRA]) with the following changes to the reaction conditions; 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
To exclude inhibition as a contributor to negative results, separate samples were spiked with an equal volume of genomic parasite DNA and run in parallel with an unspiked specimen. Control DNA extracted from an infected clinical sample was used as a positive control for the PCR assays.
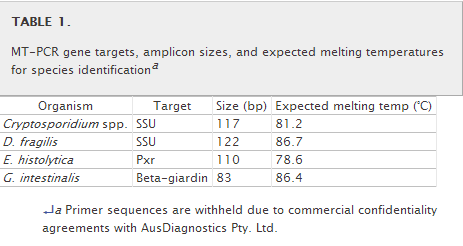
MT-PCR. (i) Primer design.To determine suitable areas for primer production, consensus sequences were generated from available GenBank sequences for each species. The sequence data from different species were then aligned with CLUSTAL_W to identify variable regions suitable for species-specific primer design (http://www.ebi.ac.uk/Tools/clustalw2/). Primers were designed from the consensus sequences with AusDiagnostics Pty. Ltd. (Alexandria, New South Wales, Australia) software. The SSU rRNA gene regions were used as targets to design specific primers for Cryptosporidium spp. and D. fragilis. The beta-giardin gene was used as a target for G. intestinalis. The pxr gene was chosen as the target for E. histolytica because it offered more sequence variation with E. dispar than other targets considered. Primer sequences are not shown due to commercial confidentiality agreements with AusDiagnostics Pty. Ltd. (Table 1). The theoretical specificities of all primer sequences were tested with the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
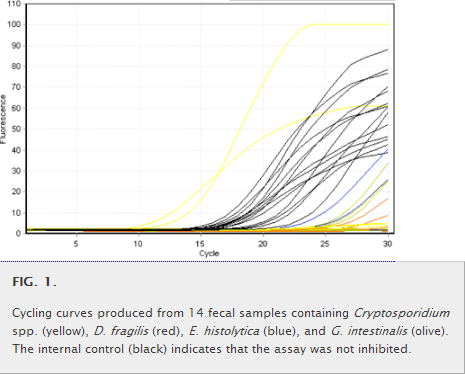
(ii) MT-PCR amplification and detection.MT-PCR is a two-step assay using nested primer pairs in which the first step involves a highly multiplexed reaction to preamplify multiple targets for between 15 and 20 cycles. These are then aliquoted into individual reaction tubes containing nested specific PCR primers as templates for the second-step reaction, which is performed using a liquid-handling robotics system provided by AusDiagnostics Pty. Ltd. (Sydney, Australia) (Fig. 1). For MT-PCR, the following items were placed on the deck of the liquid-handling system: a strip tube containing step 1 multiplexed primers was placed in the thermal cycler; a gene disc containing lyophilized step 2 primers was placed in a loading block; and oil (for covering PCR mixtures), master mix, and water tubes (all supplied in a kit form) were placed in a reagent block. The samples were directly added to the strip tube in the thermal cycler. A software template for the reaction was then selected, and all operations for performing the step 1 multiplexed preamplification, dilution, and aliquoting into the step 2 reaction tubes in the gene disc were performed automatically. The gene disc was then hermetically sealed in a heat sealer, and step 2 amplification was carried out in a Rotor-Gene RG6000 thermal cycler. At the end of step 2, the presence or absence of each target was automatically called using a software routine (AusDiagnostics Pty. Ltd.) that compared the melting temperature of the product with expected values and checked the purity and quantity against predetermined threshold values, which were all manually verified. Master mix reagents and 72-well gene discs containing lyophilized primers were prepared and supplied by AusDiagnostics Pty. Ltd.
(a) First-round multiplexed preamplification.PCRs were performed in a 20-μl volume consisting of 10 μl step 1 master mix (containing reverse transcriptase, Taq, deoxynucleoside triphosphates (dNTPs), buffer, and salts; AusDiagnostics, New South Wales, Australia) and 10 μl of template DNA plus water. For this study, 5 μl of sample extract was used. Amplification was performed for 15 cycles in the Easy-Plex liquid-handling system.
(b) Second-round quantification amplification.After completion of the step 1 cycling, a sample was automatically taken in the Easy-Plex liquid-handling system, diluted, and aliquoted into wells of a 72-place gene disc containing step 2 primers.
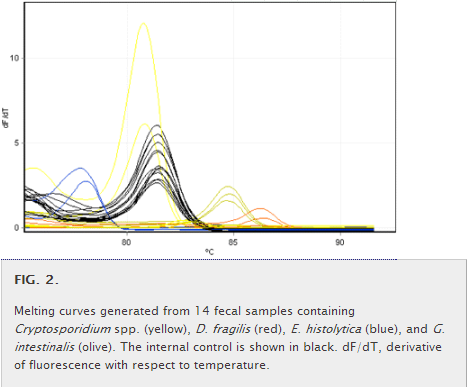
Amplification was performed on the RG6000 thermal cycler. Following cycling, a melting curve (Fig. 2) was generated from 75 to 95°C at 0.4°C intervals. The expected melting temperature of each protozoan target was established from pure-culture DNA. The presence or absence of an organism was determined by analysis software (AusDiagnostics) that compared the given melting temperature to the expected melting temperature. An internal reaction control was included for each specimen to monitor for PCR inhibition.
(iii) Control group.To determine if the MT-PCR would cross-react with stool samples containing parasites and bacteria other than the four target organisms, a control group comprising 100 fecal samples from patients and containing adenovirus (n = 5), rotavirus (n = 5), norovirus (n = 5), Campylobacter spp. (n = 5), Salmonella spp. (n = 5), Shigella spp. (n = 5), Clostridium difficile(n = 5), E. dispar (n = 20), E. moshkovskii (n = 20), Entamoeba coli (n = 5), Entamoeba hartmanni (n = 5), Blastocystis hominis (n = 5), Endolimax nana (n = 5), Iodamoeba butschlii (n = 2), Chilomastix mesnili (n = 1), Cyclospora (n = 1), and Pentatrichomonas hominis (n = 1) was included in the study. All samples were from previous studies, and the bacterial and parasite flora had been defined by a combination of microscopy, phenotypic identification, and PCR (6, 21, 23, 24). This control group underwent DNA extraction and MT-PCR as described above.
A total of 472 fecal samples submitted to the Department of Microbiology at St. Vincent's Hospital were included in the study, along with a control group of 130 patient samples. A sample was considered a true positive if it was positive by 2/3 of the diagnostic methods used. The results found in this study are summarized in Table 2.
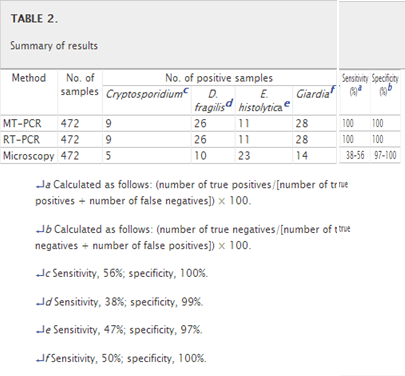
A total of 74/472 samples were positive by MT-PCR. There was 100% correlation between MT-PCR and RT-PCR. Both the MT-PCR and RT-PCR assays detected 28 cases of G. intestinalis infection, 26 cases of D. fragilis infection, 11 cases of E. histolytica infection, and 9 cases of Cryptosporidium sp. infection in the clinical samples. Both RT-PCR and MT-PCR showed 100% sensitivity and specificity. Mixed infections were detected in 4 samples. There was no evidence of inhibition of the amplification in any of the samples, with the internal control of the MT-PCR producing a product, along with the control spiked RT-PCR samples.
With microscopy, only 52/472 samples were positive for one or more of the enteric protozoa. Microscopy detected only 14 cases of G. intestinalis infection, 10 cases of D. fragilis infection, 23 cases of E. histolytica infection, and 5 cases of Cryptosporidium sp. infection in the clinical samples. However, of these, only 38 were true positives. It should be noted that microscopy cannot differentiate the nonpathogenic, morphologically identical E. dispar and E. moshkovskii from the pathogenic E. histolytica. Out of the 23 microscopy-positive E. histolytica samples, compared to the PCR methods, only 7 were true E. histolytica positives. Upon review of the iron-hematoxylin-stained slides, it was determined that 15 were morphologically identical to E. histolytica (i.e., E. dispar/E. moshkovskii), while one was determined to be the nonpathogenic Entamoeba coli. Also, one false positive was also seen in the D. fragilis microscopy-positive group, compared to the PCR assays, and on review of the permanently stained smear, it was determined that the isolate was a nonpathogenic E. nana. When microscopy was compared to both molecular methods, the sensitivities varied from 38% for D. fragilis to 47% for E. histolytica, 50% for Giardia, and 56% for Cryptosporidium, while the specificities also varied from 97% for E. histolytica to 99% for D. fragilis and 100% for Giardia and Cryptosporidium.
Out of the 100 samples in the control group, none of the samples run by MT-PCR produced a product. No cross-reactivity was seen with the following organisms: adenovirus, rotavirus, norovirus, Campylobacter spp., Salmonella spp., Shigella spp., C. difficile, E. dispar, E. moshkovskii, Entamoeba coli, E. hartmanni, B. hominis, E. nana, I. butschlii, C. mesnili, Cyclospora, and P. hominis.
E. histolytica, G. intestinalis, Cryptosporidium, and D. fragilis are the four most important and commonly occurring diarrhea-causing parasitic protozoa. It is therefore essential that correct diagnosis be made, as all four protozoa can be successfully treated with a range of antiprotozoal drugs. The MT-PCR assay for the detection of Cryptosporidium, Dientamoeba, E. histolytica, and G. intestinalis presented here provides an additional diagnostic tool for the rapid, sensitive, and specific detection of these enteric protozoa.
In both the clinical samples and control samples tested the MT-PCR for the detection of Cryptosporidium, Dientamoeba, E. histolytica, and G. intestinalis achieved a sensitivity and specificity of 100%. Compared to previously published RT-PCR assays targeting the same organisms, the MT-PCR results correlated 100% with the RT-PCR results. In all samples tested in which microscopy revealed the presence of Cryptosporidium, Dientamoeba, E. histolytica, and G. intestinalis, specific amplification was detected. However, MT-PCR detected an additional 36 positive samples (4 Cryptosporidium, 16 D. fragilis, 4 E. histolytica, and 14 Giardia). The assay also was found not to cross-react with various other viral, bacterial, and protozoal fecal pathogens.
This study highlights the lack of sensitivity that conventional staining techniques that are commonly used in most diagnostic laboratories provide for the diagnosis of these infections. Microscopy detected only 38/472 positive samples compared to 74/472 for the MT-PCR assay. Compared to both RT-PCR assays, the sensitivity of microscopy ranged from 38% for D. fragilis up to 56% for Cryptosporidium. Previous studies have produced similar findings. When comparing microscopy, culture, conventional PCR, and RT-PCR for the detection of D. fragilis, it was found that, compared to RT-PCR, microscopy had a sensitivity of only 34%; this is similar to the 38% found in this study. The implication of using only microscopy for the detection of D. fragilis is that laboratories may be missing up to 2/3 of cases. Detection of the other parasite-specific DNAs has also been shown to be more sensitive than microscopy, as it has for Giardia infections, for Cryptosporidium infections, and for amoebic infection with E. histolytica- and E. dispar-specific (real-time) PCR (29).
MT-PCR is essentially a nested RT-PCR technique. The “nested” feature of the MT-PCR contributes to the high level of sensitivity and specificity, as it depends on two different sets of specific primers for both rounds of amplification. However, the first stage of the multiplex PCR involves PCR for only 10 cycles of amplification. This prevents competition between the primers, and by diluting the first-round product 50-fold for the second-round PCR, carryover of primers from the first round is negligible. This prevents the step 1 primers from taking part in the step 2 reaction and avoids problems with PCR inhibition. The MT-PCR assay also includes an internal control, that is, a contrived sequence that does not appear in nature, to monitor for PCR inhibition. The system uses a liquid-handling robot to automate the process; this enables staff with little or no molecular experience to operate the assay. The platform also uses automated software analysis that determines the presence or absence of the organism. The software routine compares the melting temperature of the target with the expected values and checks the purity and quantity against predetermined threshold values. The other advantage of this MT-PCR assay is that up to 72 targets can be multiplexed compared with 3 to 5 with a conventional multiplex RT-PCR. As this assay utilizes intercalating dye detection, expensive probes are not required, and only a single-channel cycler is needed, which keeps hardware costs down compared to a multichannel cycler. Finally, multiplex RT-PCR assays require extensive and quite often complex optimization, while the MT-PCR does not. The assay we describe presently costs around $12.88 (Australian) ($12 U.S.) for 4 targets (approximately $3.22 [Australian] per target), including the internal control that is run with each reaction. Even with additional purification and labor costs, this is potentially a cost-effective technique for the diagnosis of the four enteric protozoa in routine diagnostic laboratories. While more expensive than an in-house RT-PCR, this MT-PCR assay has the advantage of being commercially available. Altogether, the MT-PCR assay is a rapid and cost-effective platform, requiring <3 h for the whole procedure (DNA extraction, MT-PCR, and analysis).
In the future, other tandem multiplex assays combining other parasitic targets could be developed, for example, for the detection of Cryptosporidium, Dientamoeba, E. histolytica, G. intestinalis, Cyclospora cayetanensis, Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Cystoisospora belli in patient samples. The implementation of such multiplex assays will have a tremendous impact on routine diagnostic laboratories, as these parasite targets could be combined with both viral and bacterial causes of diarrhea. This would represent a major advance in the differential laboratory diagnosis of diarrheal diseases in general.
In conclusion, the MT-PCR described here is a sensitive and specific method for the detection of Cryptosporidium, Dientamoeba, E. histolytica, and G. intestinalis. It also offers the possibility of introducing DNA detection in many laboratories where traditional multiplex RT-PCR may not be feasible, given the user-friendly platform that requires no previous molecular biology experience. This study also highlights the fact that MT-PCR and RT-PCR should be considered the gold standard and as such the method of choice for the diagnosis of enteric protozoan disease.
This work was supported by a grant from the Institute of Laboratory Science at St. Vincent's Hospital, Sydney, Australia.
▿ Published ahead of print on 3 November 2010.