发布者:Primerbank 时间:2017-09-25 浏览量:853
Alberto Nonis , Marco Scortegagna , Alessandro Nonis , Benedetto Ruperti
Department of Environmental Agronomy and Crop Productions, University of Padua, Viale dell’Università 16, 35020 Legnaro (PD), Italy
University Centre of Statistics for Biomedical Sciences, Vita-Salute San Raffaele University, Via Olgettina 58, 20132 Milano, Italy
article info
Article history: Received 19 October 2011
Available online 6 November 2011
Keywords: Gene expression ;Sequence features ;qPCR ;Primer
Abstract
An essential pre-requisite to perform sound quantitative real-time polymerase chain reaction (qPCR) assays is to design outstanding primer pairs. This means they must have a good efficiency and be not prone to produce multiple amplicons or primer dimer products. To circumvent these issues, several softwares are available to help primer design. Although satisfactory computer-aided primer design tools are available for standard PCR, less efforts were done to provide specific methods for selection of optimal primer pairs for qPCR. We have developed PRaTo a web-based tool that enables checking and ranking of primers pairs for their attitude to perform optimally and reliably when used in qPCR experiments. PRaTo is available at http://prato.daapv.unipd.it.
1. Introduction
Quantitative real-time polymerase chain reaction (qPCR) is a technique combining DNA (or cDNA) amplification by polymerase chain reaction (PCR) with real-time detection of products [1]. PCR amplifies a DNA fragment included between two short oligonucleotides (around 20 bases long), theoretically doubling the quantity of product at every cycle (when reaction efficiency is assumed to be 100%). To detect in real-time the quantity of formed products, the most popular approach uses an intercalating fluorescent dye that binds double stranded DNA. So, assay specificity is granted mainly by the primer pair, and consequently its optimal design is essential to perform reliable qPCR experiments. In principle, even though primers for qPCR are substantially not different from those for conventional PCR, they need to meet special features [1]. They should enable strictly the synthesis of a single amplicon, with good efficiency (ideally 2 copies of template after every PCR cycle) and without formation of primer dimers. Therefore, with the exception of those cases where a validated assay for the gene of interest already exists [2–4], designing good primer pairs is an essential step. Some web-based tools are offered for this purpose by companies supplying oligonucleotides, and applications are also available for free on the net [5]. However, the majority of these algorithms have been conceived for conventional PCR, although they are generally of help also for qPCR primer design. Some of them (e.g. Primer3; [6]) return a huge selection of primer pairs that are in some way ranked, but no specific criteria for qPCR are assumed. Furthermore, in specific situations, these applications can not be used and design by hand is requested. An example is when one deals with multigene families, a task that is becoming mandatory in gene expression analyses especially after the sequencing of several genomes. To discriminate between different transcripts from genes belonging to the same family, one has few choices on the primer landing sequence and only tedious and time-consuming design by hand is possible. In this case, just checking fundamental oligonucleotide properties such as Tm with specific web tools (e.g. Oligonucleotide Properties Calculator, [7]) is possible, but there are no ways to rank them and foresee which pair will give the best result in qPCR assays.
In this work a new web tool (PRaTo; phonetic: / prato/) was implemented to check and rank primer pairs for their qPCR performing potential, taking into account the qPCR primer design consideration described by Nolan et al. [1]. Moreover, as primer dimers strongly impact quality of qPCR experiments, an in depth search for primer annealing sites was included in PRaTo.
2. Materials and methods
2.1. Algorithm and website implementation
PRaTo was written in PHP language (ver. 4.4.9) and it is freely available at http://prato.daapv.unipd.it. The main page gives the user the possibility to insert up to 5 primers pairs. Only A, T, G, C letters are accepted. Once the form is filled and the ‘‘RANK’’ button has been pressed, a table will appear reporting results. Each primer pair score is indicated in the row ‘‘ALL’’. This represents the sum of the score of each primer (row ‘‘FOR’’ and ‘‘REV’’) and their interaction (row ‘‘X’’). The rules used for the implementation of PRaTo were based on the considerations on primer design, made by [1]. For the determination of primers ranking, a score was associated to each primer feature as reported in the following points:
Optimal primer length between 15 and 25 bases: Shorter primers are not accepted, longer primers received a score of 1 for each nucleotide exceeding 25 bases.
G/C content around 50%: A range between 40% and 60% was considered as acceptable (between 8 and 12 G/C for a 20 nucleotides primer). A score of 1 was applied for each 1% exceeding the acceptable range (e.g. 35% or 65% G/C were given a score of 5).
Having one G or C at 30 terminal position preceded by one A/T was considered as positive for the control of mispriming. When this feature was not found, a 1 score was assigned.
No more than 3 G or C should be present in the last five bases of 30 -end to avoid ambiguous binding. If 4 or 5 G/C were found, a score of 7 or 10 was assigned respectively.
Long runs of a single base should be avoided (especially G or C). If runs longer than 4 were present, a score of 1 was counted for each G or C, 0.75 for each A or T.
Primers should have a DG > 10 kcal/mol, since primer dimers have a negative DG value. Since DG is a mere index of primerdimer potential formation, we preferred to focus on removing all the potential annealing sites. So, primer sequences were scanned for potential annealing sites made of 4 or more nucleotides. This search was conducted for each primer against itself, and between the two forming a primer pair. A negative score was given in an amount equal to half the length of the annealing site. If the annealing site was at 30 terminus of one primer, the score was double the length of the annealing site. While, if it was at 30 terminus for both primers, score was three times the length of the annealing site. Secondary structures of primers were scored in the same way. ‘
‘ALL’’ score was rounded to the superior integer. 2.2. PRaTo va
2.2. PRaTo validation
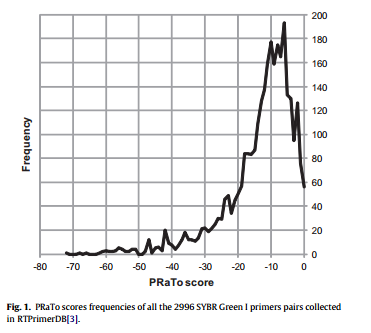
To test PRaTo functionality all the 2996 SYBR Green I primer pairs from established assays and collected in the qPCR primer database (RTPrimerDB; http://www.rtprimerdb.org; [3]) were retrieved and processed by PRaTo algorithm. In particular, ‘‘ALL’’ scores were retrieved and their frequency was discussed.
3. Results and discussion
As far as score interpretation is concerned, the closer the score of the primer pair to zero, with ‘‘0’’ being the score of a primer pair fulfilling at best all the listed requirements, the higher the chance for optimal performance in qPCR. Even though it is not possible to fix a threshold value giving certainty of results, as primers are not the only players in qPCR reactions, we tried to establish reference ranking values. To this end we analyzed with PRaTo primer pairs sequences present in RTPrimerDB repository [3].
As showed in Fig. 1, the frequency distribution of PRaTo scores pointed the maximum frequency of working primers on a score range comprised between 0 and 6. This frequency showed a gradual decrease for scores comprised between 6 and 11 and dropped drastically for scores below 11. This would suggest that the range between 0 and 6 most likely includes the best performing primer pairs, with those ranked between 7 and 11 being still acceptable. The rapid decrease of number of primer pairs with scores lower than 11, would mean that just few primer pairs are functional in this range and therefore should be discarded or considered with caution.
PRaTo is a simple to use and easy to interpret web-based tool de-signed to help scientists to rank primer pairs on the base of their qPCR performing potential. It can be used as a stand-alone tool, or in association with softwares for primer design or for calculating oligonucleotide properties. Since PRaTo score gives an estimation of the primer pair quality, we suggest inserting its score on materials and methods of papers to help interpretation of reliability of qPCR outcomes. Ac
Acknowledgments
The authors are greatly indebted to Alberto Pasimeni (University of Padua) for help with PRaTo website hosting. Alberto Nonis is supported by University of Padua (Assegno di ricerca senior).
References
[1] T. Nolan, R.E. Hands, S.A. Bustin, Quantification of mRNA using real-time RTPCR, Nat. Protoc. 1 (2006) 1559–1582.
[2] W. Cui, D.D. Taub, K. Gardner, QPrimerDepot: a primer database for quantitative real time PCR, Nucleic Acids Res. 35 (2007) D805–D809.
[3] S. Lefever, J. Vandesompele, F. Speleman, F. Pattyn, RTPrimerDB: the portal for real-time PCR primers and probes, Nucleic Acids Res. 37 (2008) D942–D945.
[4] A. Spandidos, X. Wang, H. Wang, B. Seed, PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification, Nucleic Acids Res. 38 (2010) D792–D799.
[5] K.A. Abd-Elsalam, Bioinformatic tools and guideline for PCR primer design, Afr. J. Biotechnol. 2 (2003) 91–95.
[6] S. Rozen, H.J. Skaletsky, Primer3 on the WWW for General Users and for Biologist Programmers, in: S. Krawetz, S. Misener (Eds.), Bioinformatics Methods and Protocols: Methods in Molecular Biology, Humana Press, Totowa, NJ, 2000, pp. 365–386.
[7] W.A. Kibbe, OligoCalc: an online oligonucleotide properties calculator, Nucleic Acids Res. 35 (2007) W43–W46