发布者:Primerbank 时间:2017-09-30 浏览量:2459
环状RNA研究进展
Steven P. Barrett, Julia Salzman
Development 2016 143: 1838-1847; doi: 10.1242/dev.128074
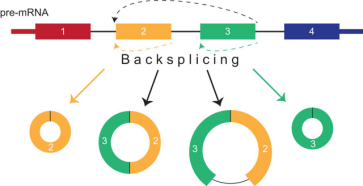
circRNA splicing and isoform diversity. A great diversity of circular RNAs (circRNAs) can be generated from a single genomic locus. These circRNAs are created through a non-canonical splicing process known as ‘backsplicing’ in which a downstream splice donor is joined to an upstream splice acceptor. Such circRNAs can consist of one or more exons and can even contain unspliced intronic sequences. Colored bars, exons; black lines, introns.
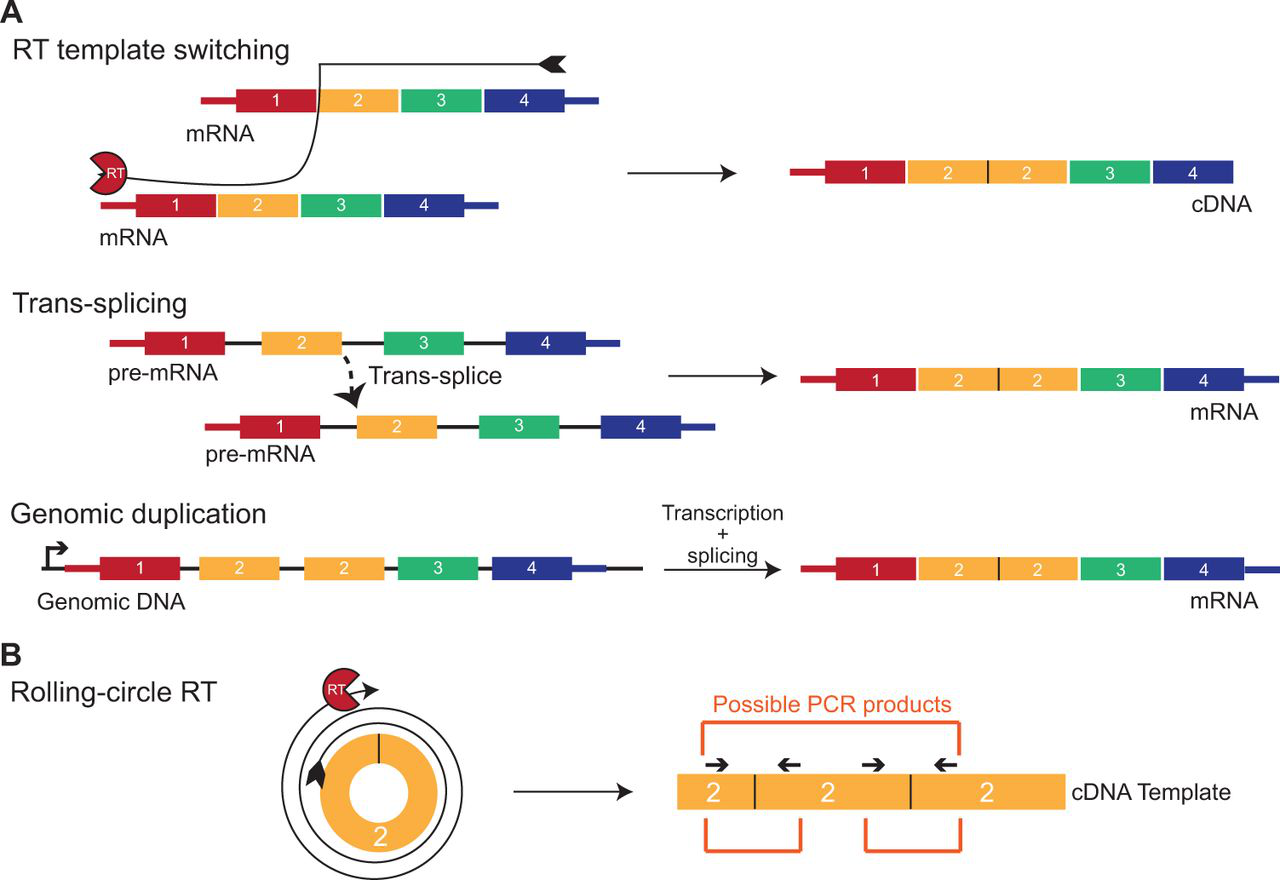
circRNA detection: sources of backsplice junctional sequences. Backsplice junctional sequences can arise from a variety of sources, resulting in false-positive detection and improper quantification of circRNAs. (A) Reverse transcriptase (RT) template switching, trans-splicing, and genomic duplication events can all give rise to linear products with spurious backsplice junctional sequences. These products can be amplified by PCR but are not derived from circRNAs. (B) Rolling-circle RT occurs when a short circRNA is converted to a long cDNA, leading to a concatamer of PCR-amplifiable sequences. Because the primers can bind to this concatemeric sequence in several locations, many PCR products can be created from a single cDNA template.
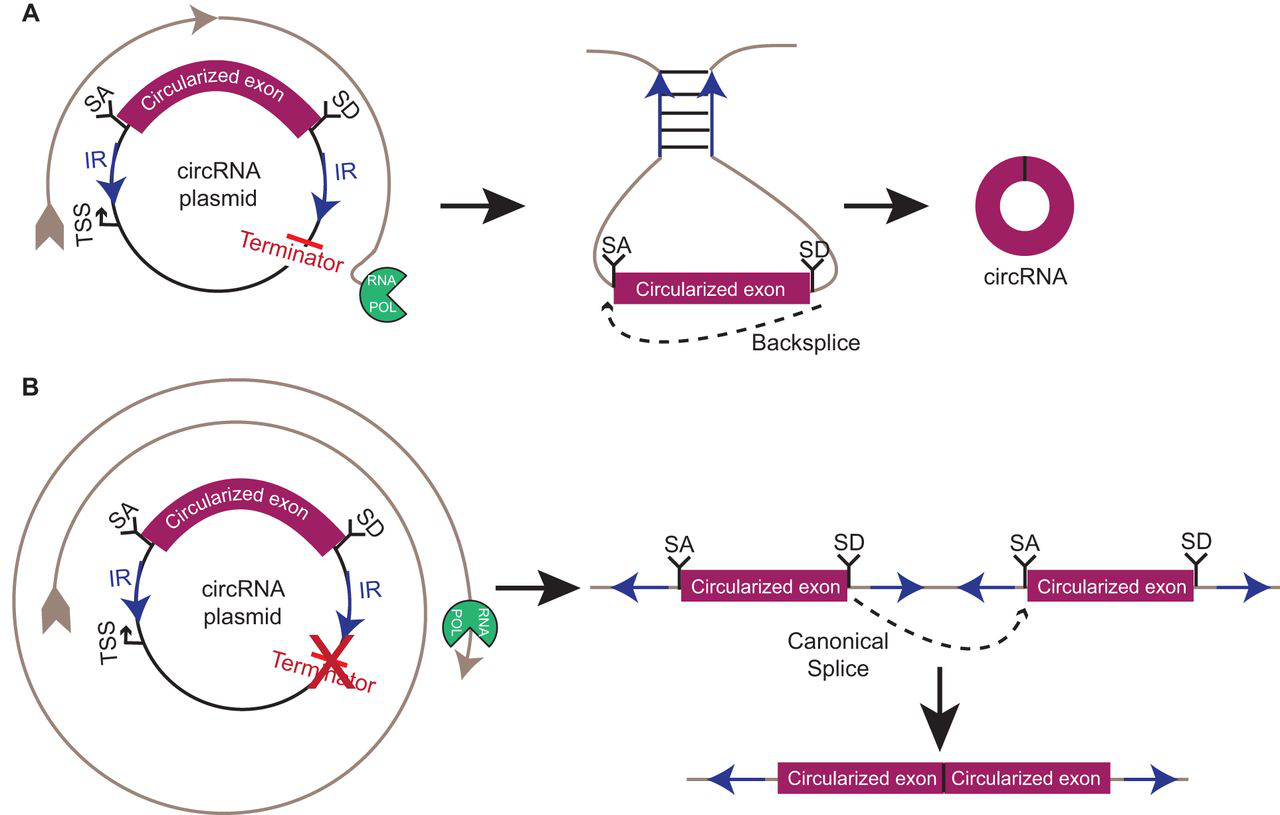
circRNA overexpression vectors. circRNA overexpression vectors are often used as convenient tools for studying the function and biogenesis of circRNAs. (A) circRNAs are commonly overexpressed on plasmids using gene fragments under the control of a strong promoter (e.g. CMV in mammalian cell lines). Circularization is induced by inverted repeats (IR) flanking the circularized exon, which presumably bring the splice acceptor (SA) and splice donor (SD) into close proximity for backsplicing. (B) In some cases, RNA polymerase (RNA POL) can bypass termination signals, resulting in rolling-circle plasmid transcription. This can give rise to a linear transcript capable of being canonically spliced, yielding a spurious backsplice junctional sequence. TSS, transcription start site.
Detecting circRNAs using bioinformatics. A chimeric read in which the 3′ end of the read maps upstream of the 5′ end with respect to the direction of transcription is the hallmark of circRNAs in RNA-seq data. In this example, the chimeric read appears to start in exon 3 and end upstream in exon 2. However, the presence of a chimeric read does not guarantee that it was derived from a circRNA (shown here and in Fig. 2A). If paired-end sequencing is employed, read 2 can inform the RNA source of the reads. For example, if read 2 maps outside the bounds of the circRNA (as defined by the chimeric read), then the read is likely to be derived from a linear species, perhaps of artifactual origin.

Regulatory factors involved in circRNA production. (A) Muscleblind (MBL, red circle) regulates the splicing of its own pre-mRNA into a circRNA (circMbl). In the presence of low amounts of MBL (left), the mbl transcript is canonically spliced to yield a translatable mRNA encoding the MBL protein. However, when MBL levels are high (right), MBL binds to the pre-mRNA and causes it to splice into a circRNA (circMbl), thereby preventing linear splicing and translation of the MBL protein. Furthermore, acting as an RBP sponge, circMbl can sequester MBL protein, lowering its free cellular concentration, thereby providing a feedback mechanism to regulate MBL levels. (B) Model for ADAR-mediated regulation of circRNA expression. ADAR family proteins (green circle) can edit and weaken RNA duplexes, decreasing the likelihood of circularization. (C) Model for QKI-mediated regulation of circRNA biogenesis. QKI (blue circle) binds to each intron flanking a circRNA and dimerizes to form a looped structure containing the exon(s). This looped structure promotes circularization of the exon(s)
circRNA functional classes. (A) circRNAs can function as a sponge containing several binding sites for a particular miRNA/RBP and can compete a miRNA/RBP away (dashed arrows) from its mRNA targets, altering gene expression. (B) Through an interaction with U1 snRNP, exon-intron circRNAs (EIciRNAs) can interact with transcription complexes at host genes to induce their transcription
